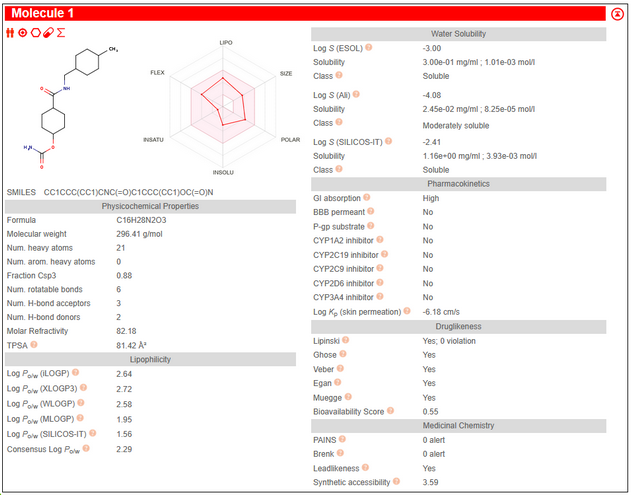
The figure above shows a typical SwissADME output. The notes below walk you through how to extract the most important information for evaluating whether a compound is suited to a particular target class.
Target-Specific Screening Ranges
| Target Class | Molecular Weight | Consensus LogP | TPSA | Why It Matters |
|---|---|---|---|---|
| Kinase | ≤ 500 Da | 2–4 | 75–120 Ų | Balances polarity for ATP pocket binding and cell entry. |
| GPCR | ≤ 450 Da | 3–5 | < 90 Ų | Small, lipophilic ligands cross membranes efficiently. |
| Protease | ≤ 550 Da | 1–3.5 | 90–140 Ų | Greater polarity helps engage catalytic residues. |
How to Work Through the Panel
-
Start by locating three fields in the Lipophilicity and Physicochemical sections:
- Molecular weight (MW)
- Consensus LogP (the agreed-upon lipophilicity value)
- TPSA (Topological Polar Surface Area)
-
Compare these three values to the table above.
✔ If all fall inside the range, the compound is well-aligned with that target class.
✖ If one is outside the range, note which one and think about whether it’s a small miss or a serious limitation. -
Next, scan the “Drug-likeness” and ADME predictions. These serve as supporting checks:
- Lipinski: Look for “Yes; 0 violation.”
- GI absorption: “High” suggests oral dosing is feasible.
- P-gp substrate: “No” avoids efflux risk (nice to have).
- CYP inhibition: “No” across major isoforms minimizes drug–drug interaction risk.
- PAINS/Brenk alerts: “0” flags avoids false positives or reactive groups.
Worked Example:
- MW = 296.41 Da (fits all classes)
- Consensus LogP = 2.29 → Kinase ✅; Protease ✅; GPCR ❌ (slightly too low)
- TPSA = 81.42 Ų → Kinase ✅; GPCR ✅; Protease ❌ (too low)
- Supporting ADME: Lipinski 0 violations, High GI absorption, 0 PAINS/Brenk alerts.
Verdict: This compound is best suited to a Kinase target, could be acceptable for a GPCR, and is not ideal for a Protease.
Quick Reference Checklist
- Identify your target class (Kinase / GPCR / Protease).
- Note down MW, Consensus LogP, TPSA and whether each is in-range.
- Summarize with a short rationale (why it fits or why it may still be acceptable).
- Optionally, include ADME notes (GI absorption, P-gp, CYP, PAINS/Brenk).